Notebooks
Premium
Trends
BioTuring
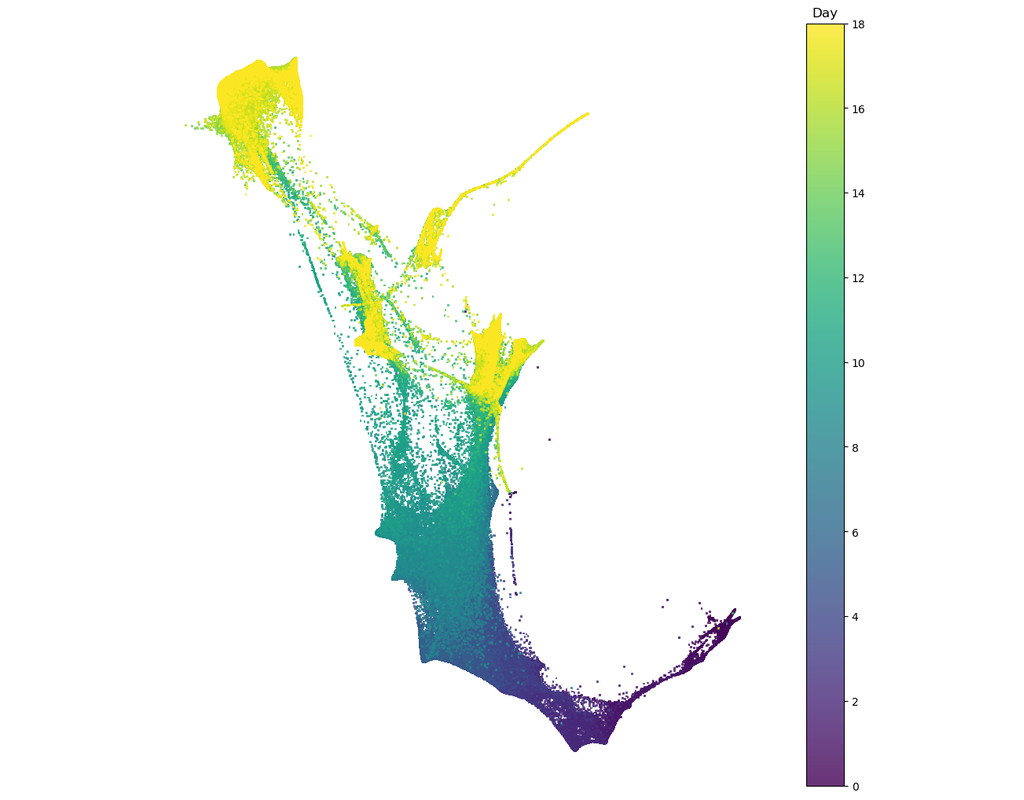
Single cell RNA-seq allows us to profile the diversity of cells along a developmental time-course. However, we cannot directly observe cellular trajectories because the measurement process is destructive. Waddington-OT is designed to infer the temporal couplings of a developmental stochastic process from samples collected independently at various time-points. The temporal couplings tell us what descendants cell x from time ti would give rise to at time tj