Notebooks
Premium
Trends
BioTuring
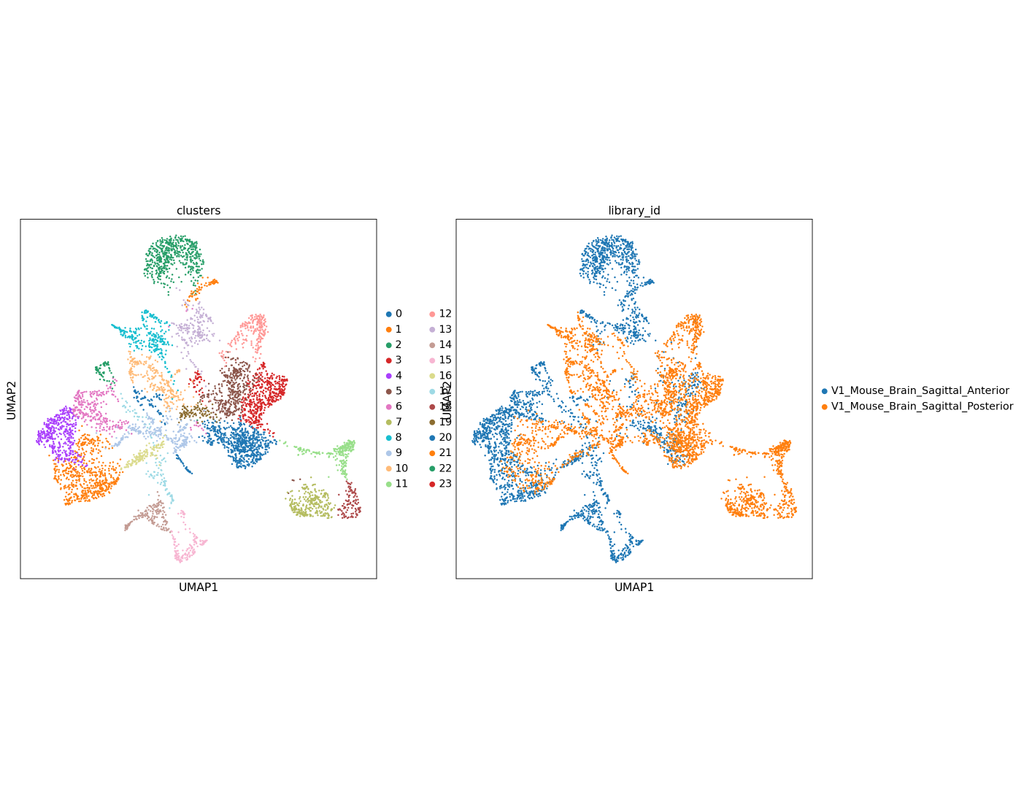
Integration of single-cell RNA sequencing (scRNA-seq) data from multiple experiments, laboratories, and technologies can uncover biological insights, but current methods for scRNA-seq data integration are limited by a requirement for datasets to derive from functionally similar cells. We present Scanorama, an algorithm that identifies and merges the shared cell types among all pairs of datasets and accurately integrates heterogeneous collections of scRNA-seq data.
Scanorama enables batch-correction and integration of heterogeneous scRNA-seq datasets, which is described in the paper "Efficient integration of heterogeneous single-cell transcriptomes using Scanorama" by Brian Hie, Bryan Bryson, and Bonnie Berger.
Scanorama is designed to be used in scRNA-seq pipelines downstream of noise-reduction methods, including those for imputation and highly-variable gene filtering. The results from Scanorama integration and batch correction can then be used as input to other tools for scRNA-seq clustering, visualization, and analysis.