Notebooks
Premium
Trends
BioTuring
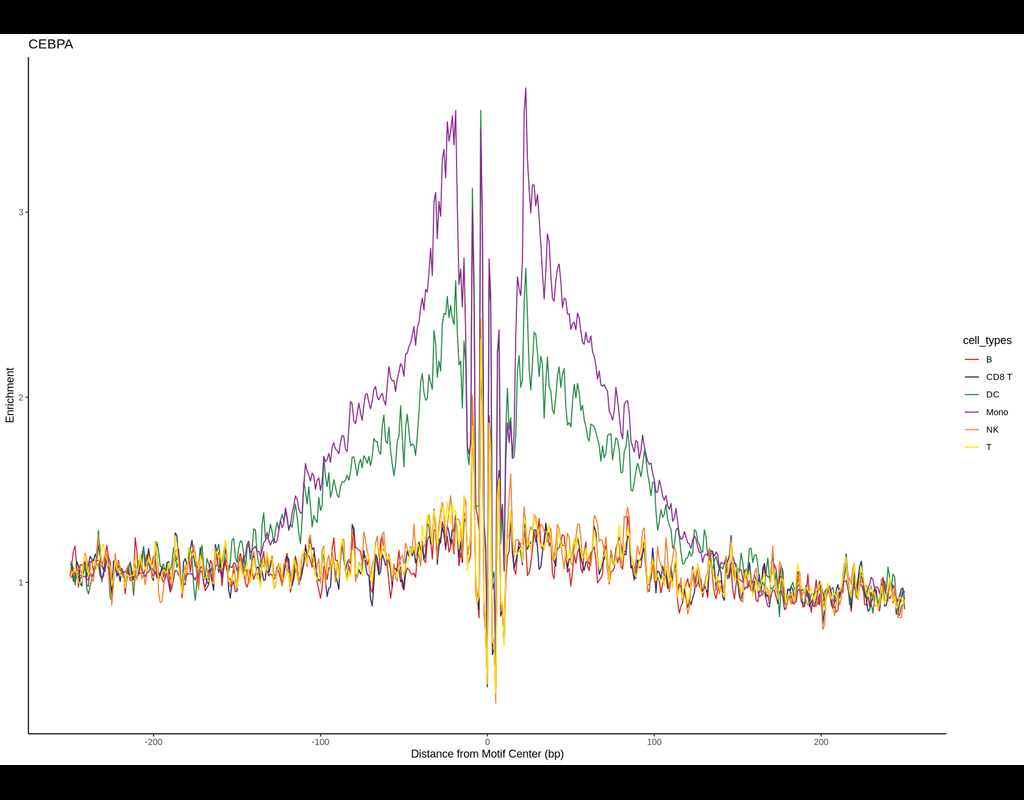
BPCells is a package for high performance single cell analysis on RNA-seq and ATAC-seq datasets. It can analyze a 1.3M cell dataset with 2GB of RAM in under 10 minutes. This makes analysis of million-cell datasets practical on a laptop.
BPCells provides:
* Efficient storage of single cell datasets via bitpacking compression
* Fast, disk-backed RNA-seq and ATAC-seq data processing powered by C++
* Downstream analysis such as marker genes, and clustering
* Interoperability with AnnData, 10x datasets, R sparse matrices, and GRanges